In this part we will see how to perform a Repartition Join in MapReduce. It is also called the Reduce-Side Join because the actual joining of data happens in the reducers. I consider this type of join to be the default and most natural way to join data in MapReduce because :
- It is a simple use of the MapReduce paradigm, using the joining keys as mapper output keys.
- It can be used for any type of join (inner, left, right, full outer …).
- It can join big datasets easily.
 We will first create a second dataset, “Projects”, which can be joined to the “Donations” data which we’ve been using since Part I. We will then pose a query that we want to solve. Then we will see how the Repartition Join works and use it to join both datasets and find the result to our query. After that we will see how to optimize our response time.
We will first create a second dataset, “Projects”, which can be joined to the “Donations” data which we’ve been using since Part I. We will then pose a query that we want to solve. Then we will see how the Repartition Join works and use it to join both datasets and find the result to our query. After that we will see how to optimize our response time.
Contents
The second dataset : Projects
The first dataset we were using, “Donations”, is a list of donations made from individuals to school projects. For example maybe an anonymous man in Austin, Texas, donated 200$ to a “Ski trip” project in a New York school. The Donations dataset has a field called “project_id” containing IDs of projects, which are actually the donation beneficiaries. This is the field we will be performing joins on. Luckily, the “projects” data is also available on the DonorsChoose Open Data.
The CSV file is around 460 MB and contains about 880k rows. You can view a sample of what its data looks like here.
As we did in Part I, we must create this following classes:
- ProjectWritable : the writable object to be used in Sequence File serialization/deserialization.
- ProjectsWriter : an executable class to parse the csv data and write it to a SequenceFile.
- ProjectsReader : an executable class to read the data (just for testing).
$ hadoop jar donors.jar data.seqfile.ProjectsWriter /data/donors/opendata_projects.csv donors/projects.seqfile 16/01/23 12:15:49 INFO zlib.ZlibFactory: Successfully loaded & initialized native-zlib library 16/01/23 12:15:49 INFO compress.CodecPool: Got brand-new compressor [.deflate] Number of lines processed : 878853 Number of errors : 0 Took 165818 ms. $ hadoop jar donors.jar data.seqfile.ProjectsReader donors/projects.seqfile 16/01/23 12:22:22 INFO zlib.ZlibFactory: Successfully loaded & initialized native-zlib library 16/01/23 12:22:22 INFO compress.CodecPool: Got brand-new decompressor [.deflate] 16/01/23 12:22:23 INFO compress.CodecPool: Got brand-new decompressor [.deflate] 16/01/23 12:22:23 INFO compress.CodecPool: Got brand-new decompressor [.deflate] 16/01/23 12:22:23 INFO compress.CodecPool: Got brand-new decompressor [.deflate] Compressed ? true 7342bd01a2a7725ce033a179d22e382d|school=9e72d6f2f1e9367b578b6479aa5852b7|total_amount=279.27|status=completed b56b502d25666e29550d107bf7e17910|school=4a06a328dd87bd29892d73310052f45f|total_amount=152.44|status=completed 3a88a47f97bd0c9b7c6a745bcd831ce3|school=66d08b506d2f3c30dec9e6fdb03cc279|total_amount=2237.8|status=completed
The final Sequence File, projects.seqfile, after keeping only 30 of the 43 fields, is around 123 MB (compressed), which conveniently fits into 1 HDFS block.
By creating the sequence file without compression, the size is about 365 MB (needing 2 blocks), but we won’t be using that. It’s good to know how big the data really is though.
The Query
Let’s try to find the results to the following query :
View all pairs of corresponding donations and projects. Each time a donation was given to a project, display the following data :
- Donation attributes : id, project id, donor city, date, total
- Project attributes : id, school city, poverty level, primary subject
This could be formulated in SQL as :
SELECT d.donation_id, d.project_id as join_fk, d.donor_city, d.ddate, d.total, p.project_id, p.school_city, p.poverty_level, p.primary_focus_subject FROM donations as d INNER JOIN projects as p ON d.project_id = p.project_id;
Our query is using an Inner Join, but we will start by using the Full (Outer) Join in the next section, for the sake of covering all use-cases.
The Repartition Join
Basic Idea and Example
The Repartition Join is a simple technique. To join our Donations with Projects, it can be described in a few easy steps :
- Creating a DonationsMapper class for the Donations data, which outputs (project_id, DonationWritable).
- Creating a ProjectsMapper class for the Projects data, which outputs (project_id, ProjectWritable).
- Receive all DonationWritable and ProjectWritable objects with the same project_id joining key and join them.
Below is an illustration of how we will join the Donations data with the Projects data, using a full outer join. For the sake of simplicity and readability, the donations sequence file only has 2 input splits (instead of 4 in reality), and the input keys are simplified (‘donXX’ for donations and ‘proXX’ for projects).
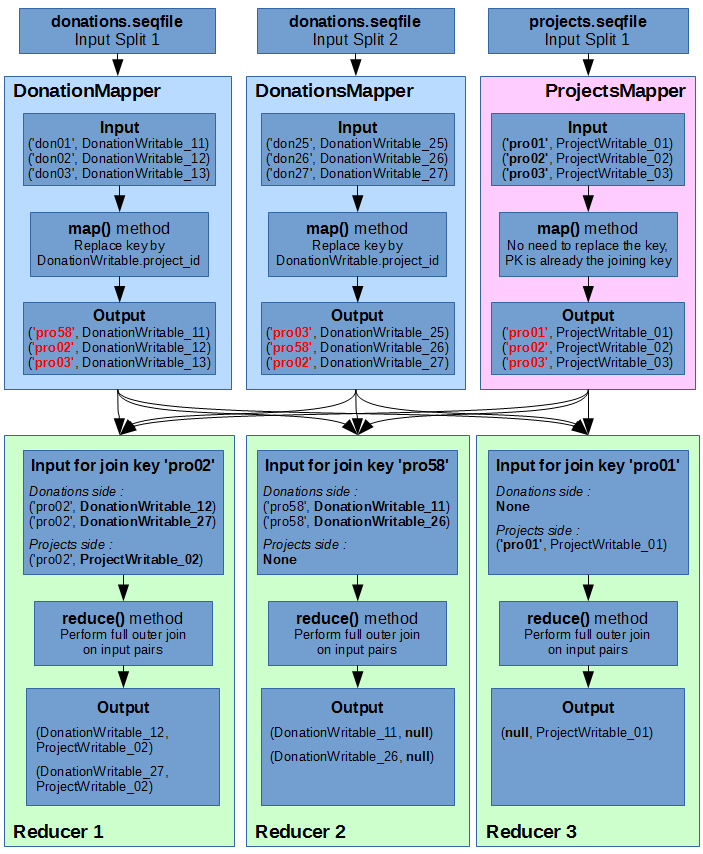
As you can see, I have illustrated 3 different cases of reduce outcomes :
- Reducer 1: Both Donations and Projects objects share the same joining key. This results in a Cartesian product of all inputs.
- Reducer 2: There is no “right side” data for the join. No projects correspond to the donations’ foreign key.
- Reducer 3: There is no “left side” data for the join. No donations correspond to the project’s primary key.
Note that the outputs of reducer 2 and 3 would be empty if it was an Inner Join.
In this illustration the outputs are the full Writable objects, which could be written to a Sequence File. But in the reduce() method we can of course decide to write out only a few attributes in a text file instead. In the following implementation we will use the latter, in order to have a human-readable output for a simpler result analysis.
Basic Implementation
Here is the code of for a basic Repartition Join between Donations and Projects: RepartitionJoinBasic.java
This job is very close to the previous illustration. It performs a Full Outer Join between DonationWritable and ProjectWritable values which share the same joining key in a given reducer. However, instead of writing the binary objects to the output, it prints out a few selected attributes (those previously specified in our query) to a plain text output.
The first thing to notice in this code is how to use 2 different reducers for our 2 different input files. This can be done very easily with Hadoop’s native MultipleInputs helper class in our driver code :
// Input parameters Path donationsPath = new Path(args[0]); Path projectsPath = new Path(args[1]); Path outputPath = new Path(args[2]); // Mappers configuration MultipleInputs.addInputPath(job, donationsPath, SequenceFileInputFormat.class, DonationsMapper.class); MultipleInputs.addInputPath(job, projectsPath, SequenceFileInputFormat.class, ProjectsMapper.class); job.setMapOutputKeyClass(Text.class); job.setMapOutputValueClass(ObjectWritable.class);
Also, since both mappers have different output types (DonationWritable for DonationsMapper and ProjectWritable for ProjectsMapper), we need to use a “generic” class which can encapsulate instances of both classes in order to receive both object types in the reducer. For this we use the ObjectWritable class from the Hadoop library. To accomplish this, we must :
- Use the ObjectWritable class as the output value class for both mapper classes, and call job.setMapOutputValueClass(ObjectWritable.class) in the driver code.
- Use objectWritable.set(donationInstance) in DonationsMapper for the mapper value output.
- Use objectWritable.set(projectInstance) in ProjectsMapper for the mapper value output.
- Use the ObjectWritable class as the input value class of the reducer class to accept inputs from both mappers.
- In the reduce() function, use objectWritable.get() to obtain the encapsulated object, which is either a donation or a project.
- Evaluate object instanceof DonationWritable to know if it is a donation. If not, it is a project.
The rest of the reduce() code is quite simple. We store all donations in a list, and all projects in another list. A this point, all elements of both lists have the same joining key, project_id, since they are all from the same call to the reduce() function. In this case, using a Full Outer Join, there are 3 cases to manage, based on the emptiness of these two lists, which I have pointed out with comments in the code. These 3 cases are also the 3 cases from Reducers 1, 2 and 3 illustrated in the big diagram from the previous section.
Job Execution and Results
I executed this job on my mini-cluster of 1 master and 4 slaves with 2 GB of RAM each, using 1 GB containers for mappers and reducers :
$ hadoop jar donors.jar mapreduce.join.repartition.RepartitionJoinBasic donors/donations.seqfile donors/projects.seqfile donors/output_repartitionjoinbasic $ hdfs dfs -ls -h donors/output_repartitionjoinbasic Found 4 items -rw-r--r-- 2 hduser supergroup 0 2016-01-29 16:16 donors/output_repartitionjoinbasic/_SUCCESS -rw-r--r-- 2 hduser supergroup 256.3 M 2016-01-29 16:16 donors/output_repartitionjoinbasic/part-r-00000 -rw-r--r-- 2 hduser supergroup 258.6 M 2016-01-29 16:15 donors/output_repartitionjoinbasic/part-r-00001 -rw-r--r-- 2 hduser supergroup 255.6 M 2016-01-29 16:16 donors/output_repartitionjoinbasic/part-r-00002 $ hdfs dfs -cat donors/output_repartitionjoinbasic/part-r-00000 ... d9e8c2cd4228fd1299588c46c7c3107e|021e635d3ba92bea0b3d59cb38cf3f13||2009-02-23 05:47:17.008|334.12 021e635d3ba92bea0b3d59cb38cf3f13|New Orleans|highest poverty|History & Geography eca0ed70d2fe84f42b235a96d47847ba|021e9d59282791705aa2e04ab6915acd||2011-03-20 20:47:53.12|100.00 null|null|null|null null|null|null|null|null 021ef537f507a68edf3458941bad28d4|Parma|highest poverty|Applied Sciences
We can see from the excerpt of the first output file that there are 3 types of joined results : perfect match, left part missing, and right part missing. In the first result from the excerpt, we can see that the donation.project_id we printed in he left side is the same as the project.project_id we printed in the right side, which is normal since they are the joining key. When either side of the join is missing, the attributes are displayed as ‘null’ strings.
The job took around 5 min 40s on average.
Optimizations
In this section we will see how to optimize the execution time of our Repartition Join. We will focus primarily on data projection, which consists in discarding all data which we don’t need before the shuffle phase.
In the Full Outer Join implementation from the previous section, the mappers wrote the “full row” objects DonationWritable and ProjectWritable to the output. And then the reducer received these full rows only to extract 4 or 5 attributes from each object. It’s a bit of waste to transfer 30 attributes only to use 5 of them. This shuffles a lot more data than we actually need.
Projection using smaller Writable objects
The first solution to this problem is to create smaller Writable classes which only contains the attributes that we need.
To reduce the amount of shuffled data from the “donations” and “projects” dataset, we will use these classes :
Then all we have to do in our DonationsMapper is to create an instance of DonationProjection for each DonationWritable full row object. And set this new instance in the generic ObjectWritable to be written in the mapper’s output. The same logic applies for the ProjectsMapper.
Here is the code for the Repartition Join job using this optimization: RepartitionJoinProjectedByWritables.java
$ hadoop jar donors.jar mapreduce.join.repartition.RepartitionJoinProjectedByWritables donors/donations.seqfile donors/projects.seqfile donors/output_repartitionjoinprojectedwritables $ hdfs dfs -ls -h donors/output_repartitionjoinprojectedwritables Found 4 items -rw-r--r-- 2 hduser supergroup 0 2016-01-29 16:58 donors/output_repartitionjoinprojectedwritables/_SUCCESS -rw-r--r-- 2 hduser supergroup 252.1 M 2016-01-29 16:58 donors/output_repartitionjoinprojectedwritables/part-r-00000 -rw-r--r-- 2 hduser supergroup 254.4 M 2016-01-29 16:57 donors/output_repartitionjoinprojectedwritables/part-r-00001 -rw-r--r-- 2 hduser supergroup 251.4 M 2016-01-29 16:58 donors/output_repartitionjoinprojectedwritables/part-r-00002 $ hdfs dfs -cat donors/output_repartitionjoinprojectedwritables/part-r-00000 | head e733b4c31f1054b1a5518ecc4e476ab8|0000023f507999464aa2b78875b7e5d6|2010-08-14 09:07:12.312|New Orleans|5.00 0000023f507999464aa2b78875b7e5d6|New Orleans|highest poverty|Health & Life Science ee519de971e6ec8961f5efa06c6ed191|0000023f507999464aa2b78875b7e5d6|2010-08-18 19:29:18.935|Fort Myers|69.26 0000023f507999464aa2b78875b7e5d6|New Orleans|highest poverty|Health & Life Science f26a3073fbec0439adbe6f2f92dc7816|0000023f507999464aa2b78875b7e5d6|2010-08-18 18:53:28.085||4.83 0000023f507999464aa2b78875b7e5d6|New Orleans|highest poverty|Health & Life Science ee7ce86ae0b07d1efb1432ad8cf2ac1c|0000023f507999464aa2b78875b7e5d6|2010-08-10 12:27:17.693||25.00 0000023f507999464aa2b78875b7e5d6|New Orleans|highest poverty|Health & Life Science
The output files are slightly smaller that before because we did an Inner Join. The few join results which didn’t have a matching left or right side were discarded. As a result, there are no “null” values in the output this time.
Using this optimization, the job took an average of 4 min 30 s.
Projection using plain text
Another solution is to manually serialize the values as plain text in the mappers. Instead of using the generic ObjectWritable for serialization/deserialization between mappers and reducers, we use simple Text writables containing values separated by a token (here I chose to use a pipe character “|”). We also need to define a special indicator somewhere in the string to define whether it is a donation or a project record. We will simply prepend the character “D” for donations and “P” for projects.
For example instead of writing a ProjectWritable to its output, the ProjectsMapper will write a line which looks like this:
D|021ef537f507a68edf3458941bad28d4|Parma|highest poverty|Applied Sciences
Then in the reducer, we can read the first character to know if it is a donation or a project, and then strip it out. By doing this we end up with the same data we had in our previous implementations.
The code for the same Repartition Join but using projection by Text values can be viewed here: RepartitionJoinProjectedByText.java
$ hadoop jar donors.jar mapreduce.join.repartition.RepartitionJoinProjectedByText donors/donations.seqfile donors/projects.seqfile donors/output_repartitionjoinprojectedtext $ hdfs dfs -ls -h donors/output_repartitionjoinprojectedtext -rw-r--r-- 2 hduser supergroup 0 2016-01-29 16:38 donors/output_repartitionjoinprojectedtext/_SUCCESS -rw-r--r-- 2 hduser supergroup 252.1 M 2016-01-29 16:38 donors/output_repartitionjoinprojectedtext/part-r-00000 -rw-r--r-- 2 hduser supergroup 254.4 M 2016-01-29 16:38 donors/output_repartitionjoinprojectedtext/part-r-00001 -rw-r--r-- 2 hduser supergroup 251.4 M 2016-01-29 16:38 donors/output_repartitionjoinprojectedtext/part-r-00002
Using this optimization, the results are exactly the same. But the job took an average of only 3 min 40 s.
It is a lot faster using text, probably because there is no serialization/deserialization done between the mappers and reducers. However we are not using objects anymore, so this technique is more prone to errors which could be caused by data stucture changes, since we are working with strings only.
Using a Bloom Filter to optimize filtering
In this post, we joined the entire datasets, without filtering any records, and saw how to perform any type of join.
However, if we need to perform some filtering on records before an Inner Join, using a Bloom Filter can significantly improve performance.
In a future post, we will explore how to use Bloom Filters in Hadoop, and use it to optimize our Repartition Join.
thanks for sharing